Menu
|
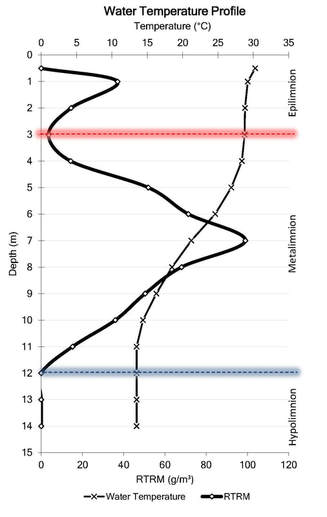
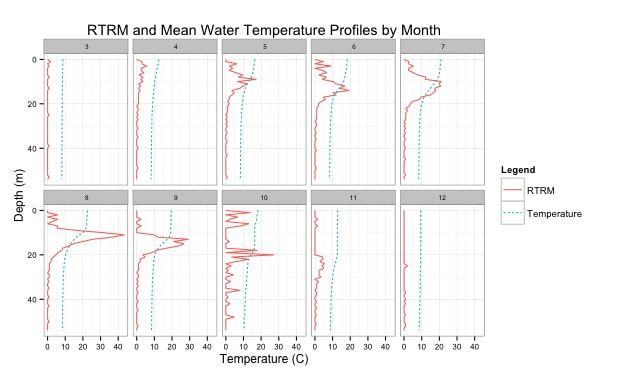
Relative thermal resistance to mixing is able to quantify stratification as a function of temperature differential, exact position of the thermocline, the exact width of the metalimnion, and how stable of stratification results from an non linear change. RTRM identifies a relatively easy computation. Instead of visually trying to identify stratification this method can be utilized to identify both the location and intensity of thermal stratification. #https://github.com/EvanAquatic #Function to calculate water density (mg/m^3) from water temperature (celsius) WaterDensityCalc <- function(x) { Density=(1000*(1-((Temperature+288.9414)/(508929.2*(Temperature+68.12963)))*(Temperature-3.9863)^2)/1000) #output is water density in mg/m^3 } Depth=Profile$Sample.Depth Temperature=Profile$Temperature Water_Density<-WaterDensityCalc(Temperature) ###Calculating water density from the defined function RTRM<-data.frame(Profile$Sampling.Location, Depth, Temperature, Water_Density) ###Creating dataframe with outputs ##Calculate RTRM RTRM$RTRM_OUTPUT= NA for(i in seq(1, nrow(RTRM))){ RTRM[i,"RTRM_OUTPUT"] = RTRM[i,"RTRM_OUTPUT"] = ((RTRM[i+1,"Water_Density"] - RTRM[i,"Water_Density"])/(1-0.9999919)) } #1000*(1-((4+288.9414)/(508929.2*(4+68.12963)))*(4-3.9863)^2)/1000=1 #1000*(1-((5+288.9414)/(508929.2*(5+68.12963)))*(5-3.9863)^2)/1000=0.9999919 RTRM$RTRM_OUTPUT[RTRM$RTRM_OUTPUT<0]<-0 #Removing negative values RTRM #######General Plots and Figures library(ggplot2) library(MASS) library(reshape) library(reshape2) #Incorporating influences of month or date #RTRM$NewDate <- format(as.Date(RTRM$RTRM1.Date, format="%m/%d/%Y"), "%Y/%m") #RTRM$Monthday <- format(as.Date(RTRM$RTRM1.Date, format="%m/%d/%Y"), "%m/%d") ################### #Creating new dataframes to plot water temperature and RTRM df<-data.frame(RTRM$Profile.Sampling.Location, RTRM$Depth, RTRM$Temperature, RTRM$RTRM_OUTPUT) names(df)<-c("SampleLocation", "Depth", "Temperature", "RTRM") #New datadrame output with new header names; length=lt and weight=wt newdf<- melt(df, id = c("SampleLocation","Depth")) names(newdf)<-c("SampleLocation", "Depth", "Legend", "Output") #New datadrame output with new header names; length=lt and weight=wt ###Water Temperature and RTRM Profile Plot Outputs g<-ggplot(newdf, aes(x=Output, y =Depth, color=Legend))+ theme_bw() + geom_path(aes(linetype=Legend),linejoin = 'round', size=1 ) + facet_wrap(~SampleLocation, ncol=5)+ # facet_grid(Collection.Date.ymd~Collection.Year)+ labs(title='4/26/2018 Reservoir Relative Thermal Resistance to Mixing (RTRM) and Water Temperature Profiles')+ xlab("Temperature (C) and RTRM (g/m^3)")+ ylab("Depth (m)")+ scale_x_continuous(limits=c(0,30), expand=c(0,0))+ scale_y_reverse(breaks=seq(12,0, by=-1), limits=c(12,0), expand=c(0,0)) + theme(strip.text = element_text(size=6.5, lineheight=0.1, hjust=0.5), axis.text.y = element_text(size=8, colour="black"), axis.text.x = element_text(size=8, colour="black"), panel.spacing = unit(1, 'lines'), legend.position = "top", legend.background = element_rect(color = "black", size = .1), legend.direction = "horizontal") ################### #Creating new dataframes to plot Dissolved Oxygen, pH, % Dissolved Oxygen, etc. df2<-data.frame(Profile$Sampling.Location, Profile$Sample.Depth, Profile$Dissolved.Oxygen) names(df2)<-c("SampleLocation", "Depth", "Dissolved Oxygen") #New datadrame output with new header names; length=lt and weight=wt newdf2<- melt(df2, id = c("SampleLocation","Depth")) names(newdf2)<-c("SampleLocation", "Depth", "Legend", "Output") #New datadrame output with new header names; length=lt and weight=wt ###Dissolved Oxygen Profile Plot Outputs g<-ggplot(newdf2, aes(x=Output, y =Depth, color=Legend))+ theme_bw() + geom_path(aes(linetype=Legend), size=1 ) + facet_wrap(~SampleLocation, ncol=5)+ # facet_grid(Collection.Date.ymd~Collection.Year)+ labs(title='4/26/2018 Reservoir Dissolved Oxygen Profiles')+ xlab("Dissolved Oxygen (mg/L)")+ ylab("Depth (m)")+ scale_x_continuous(limits=c(0,12), expand=c(0,0))+ scale_y_reverse(breaks=seq(12,0, by=-1), limits=c(12,0), expand=c(0,0)) + theme(strip.text = element_text(size=6.5, lineheight=0.1, hjust=0.5), axis.text.y = element_text(size=8, colour="black"), axis.text.x = element_text(size=8, colour="black"), panel.spacing = unit(1, 'lines'), legend.position = "top", legend.background = element_rect(color = "black", size = .5), legend.direction = "horizontal") [1] Kortmann, R.W. 1990. Thermal Stratification in Reservoirs: Causes, Consequences, Management Techniques. Proceedings AWWA-WQTC, San Diego, November 1990.
1 Comment
|
Archives
February 2020
Categories |
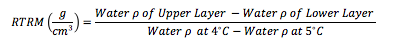
 RSS Feed
RSS Feed
